
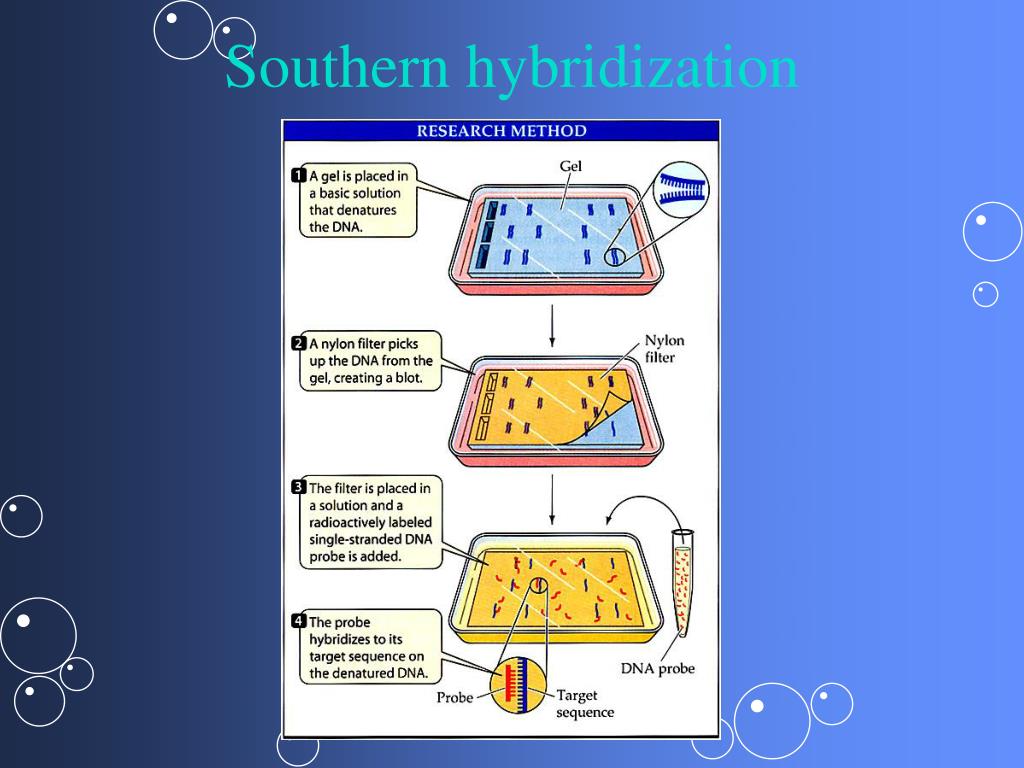
Nylon membranes are more commonly used because of the hazardous nature of baking nitrocellulose membranes in an oven at 80 ☌. The fragments are then immobilized on the membrane by UV irradiation (for nylon) or heat application (for nitrocellulose). After denaturing the fragments to yield single strands, the DNA is transferred to a nylon or nitrocellulose membrane. In Southern blotting, the target DNA is cut into smaller fragments and run on an agarose gel. This process is called Southern blotting. However, additional steps are needed to verify the sequence identity of the sample DNA fragments.ĭenatured DNA fragments must be transferred onto a carrier membrane from the gel to make it accessible to a probe - a small ssDNA fragment complementary to the target DNA and labeled with a reporter tag. Running a DNA ladder containing fragments of the known length alongside the sample helps determine the approximate length of the sample DNA fragments. Because the labeled probes will only bind to the DNA sequence of interest, any visible band indicates presence of the target DNA.Īgarose gel electrophoresis is very useful in separating DNA fragments by size. When the membrane is soaked in a buffer solution containing the probes and warmed at 42 ☌, the target ssDNA pairs with the complementary probe DNA to form a labeled, hybrid, double-stranded DNA.Īfter overnight hybridization, the membrane is washed to remove the unbound probes.įor radioactively labeled DNA, the membrane is exposed to an X-ray film for detection.Įnzyme-labeled probes can be detected by adding an appropriate substrate and visualizing the bands as the color or luminescence develops. For visualization, the probes are either labeled with a radioactive phosphorus -P-32, or an enzyme that generates an easily detectable product. The probes are short, single-stranded DNA that have a complementary sequence to the target DNA fragments. The solution coats the membrane and prevents any non-specific binding with the probe DNA. The membrane is then soaked in a solution of denatured salmon sperm DNA. This prevents diffusion and immobilizes the DNA bands. When irradiated with UV rays, the DNA becomes covalently cross-linked to the membrane. The ssDNA transfers onto the membrane through capillary action, while the high salt concentration in the neutralizing solution helps the DNA to bind. Next, a nylon membrane is placed on top of the gel and weighed down with a stack of paper towels. The gel is then placed on a sponge in a DNA neutralizing solution containing sodium chloride and tris buffer, to reset its pH to 7.0. The procedure follows five main steps: electrophoresis, denaturation, membrane transfer, hybridization, and visualization.įirst, the genomic DNA is digested by restriction enzymes, and the resulting DNA fragments are loaded on an agarose gel and separated using gel electrophoresis.įor denaturation or separation of the double-stranded DNA into two single-stranded DNA, or ssDNA, the gel is soaked in a sodium hydroxide solution. The technique is named after Edwin Southern, the British biologist who first developed it. Southern blotting is a technique where a labeled DNA probe hybridizes with a target DNA to detect a particular sequence within a genome.


 0 kommentar(er)
0 kommentar(er)
